
Supplementary material |
| Global analysis Species tree Alignments Tables SG_I SG_II SG_III SG_IV SG_V SG_VI SG_VIIa SG_VIIb SG_VIII-1 SG_VIII-2 SG_IX SG_Xa SG_Xb SG_XI SG_XIIa SG_XIIb SG_XIIIa SG_XIIIb SG_XIV SG_XV |
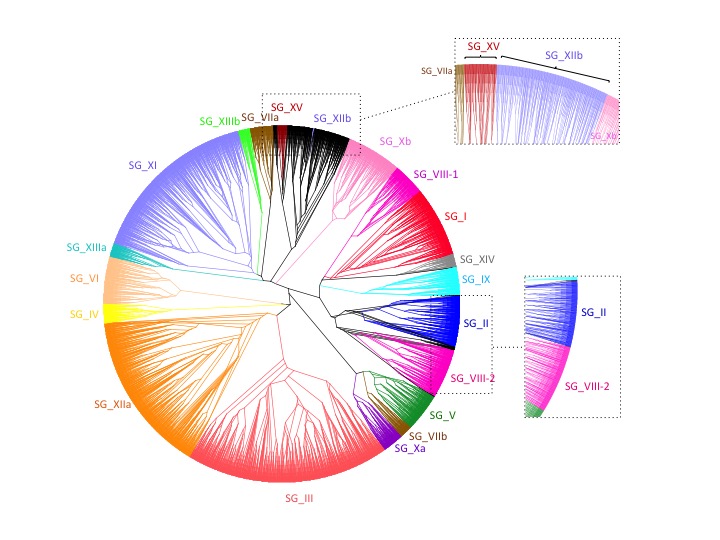
Global phylogeny is inferred using MAFFT (progressive method) then TrimAl for multiple alignment, and Protdist (JTT) then FastME (SPR) for distance phylogeny.
How to display the tree:
Can be displayed or hidden in non-interactive image clicking here.
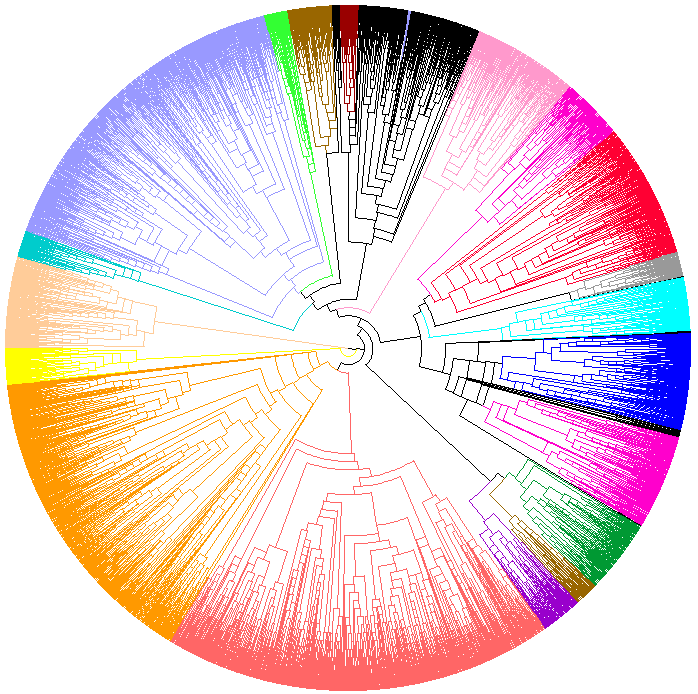
Can be displayed, fully colored, using Dendroscope and loading this file into the software.
|
Command lines used to obtain this phylogeny are:
mafft --retree 10 in.fasta > out.fasta
|